from matplotlib import pyplot as plt
import numpy as np
import pandas as pd
import geopandas as gpd
np.random.seed(42)Week 11: Clustering Analysis in Python
- Section 401
- Nov 13, 2023
Clustering in Python
- Both spatial and non-spatial datasets
- Two new techniques:
- Non-spatial: K-means
- Spatial: DBSCAN
- Two labs/exercises this week:
- Grouping Philadelphia neighborhoods by AirBnb listings
- Identifying clusters in taxi rides in NYC
“Machine learning” and “AI”

- The computer learns patterns and properties of an input data set without the user specifying them beforehand
- Can be both supervised and unsupervised
Supervised
- Example: classification
- Given a training set of labeled data, learn to assign labels to new data
Unsupervised
- Example: clustering
- Identify structure / clusters in data without any prior knowledge
Machine learning in Python: scikit-learn
- State-of-the-art machine learning in Python
- Easy to use, lots of functionality
Clustering is just one (of many) features
https://scikit-learn.org/stable/
We will focus on clustering algorithms today and discuss a few other machine learning techniques in the next two weeks. If there is a specific scikit-learn use case we won’t cover, I’m open to ideas for incorporating it as part of the final project.
Part 1: Non-spatial clustering
The goal
Partition a dataset into groups that have a similar set of attributes, or features, within the group and a dissimilar set of features between groups.
Minimize the intra-cluster variance and maximize the inter-cluster variance of features.
Some intuition
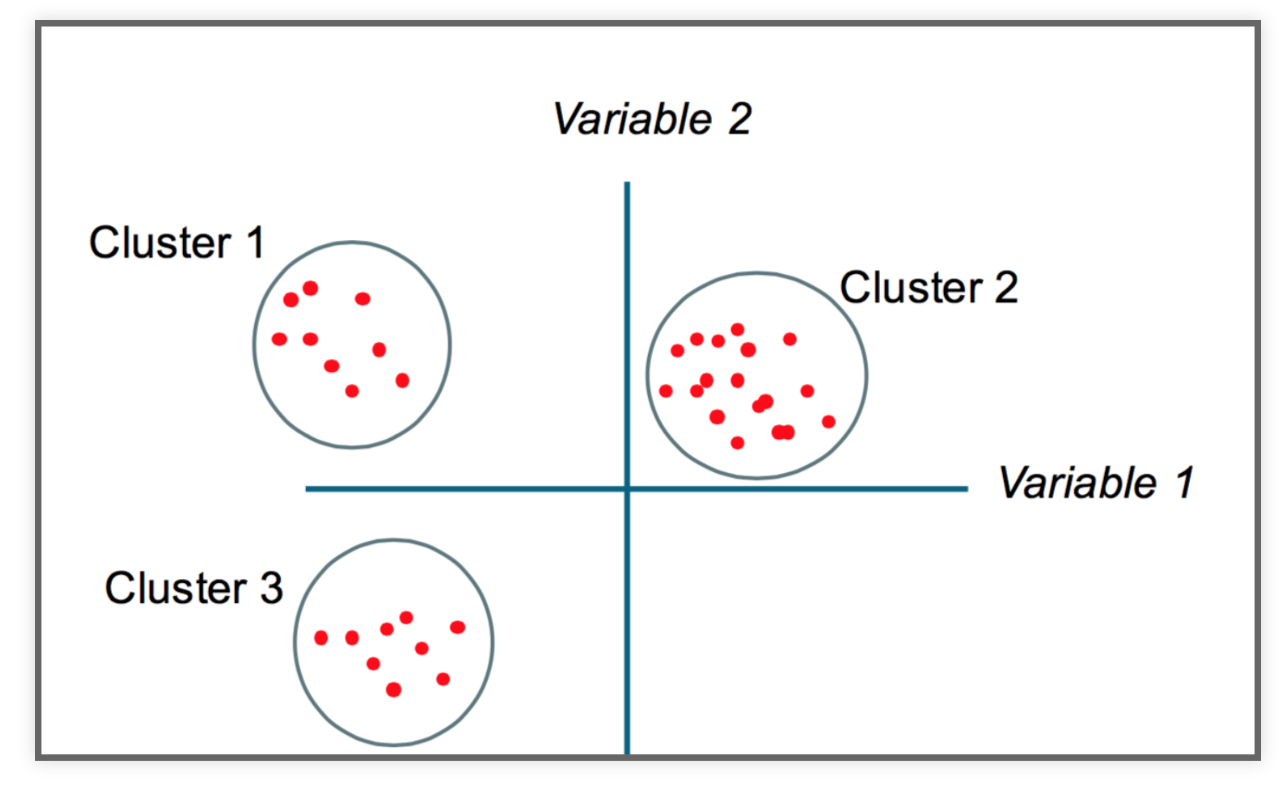
K-Means clustering
- Simple but robust clustering algorithm
- Widely used
- Important: user must specify the number of clusters
- Cannot be used to find density-based clusters
This is just one of several clustering methods
https://scikit-learn.org/stable/modules/clustering.html#overview-of-clustering-methods
Check out Andrew Ng’s Coursera lecture on unsupervised clustering.
How does it work?
Minimizes the intra-cluster variance: minimizes the sum of the squared distances between all points in a cluster and the cluster centroid
K-means in action
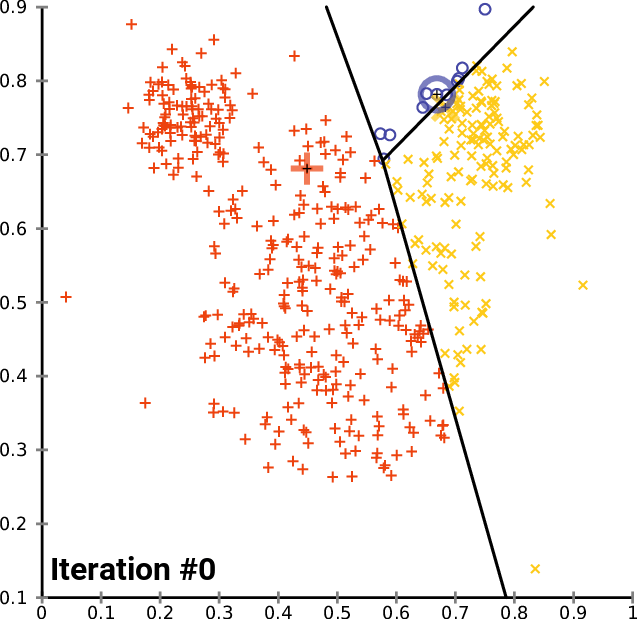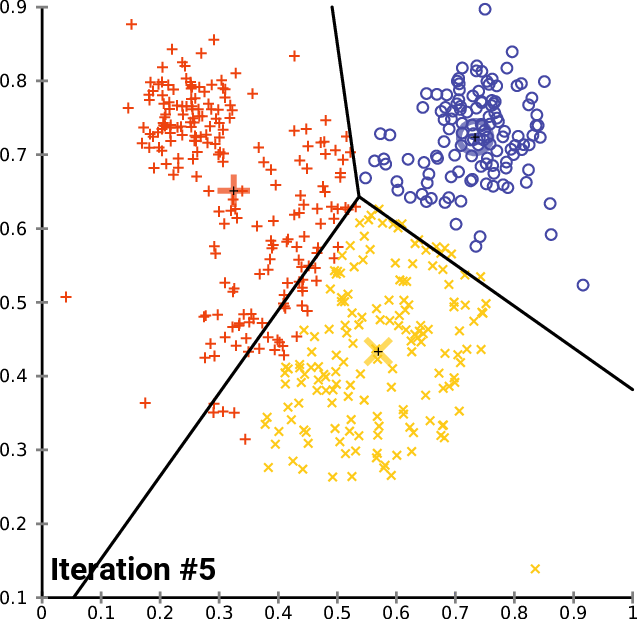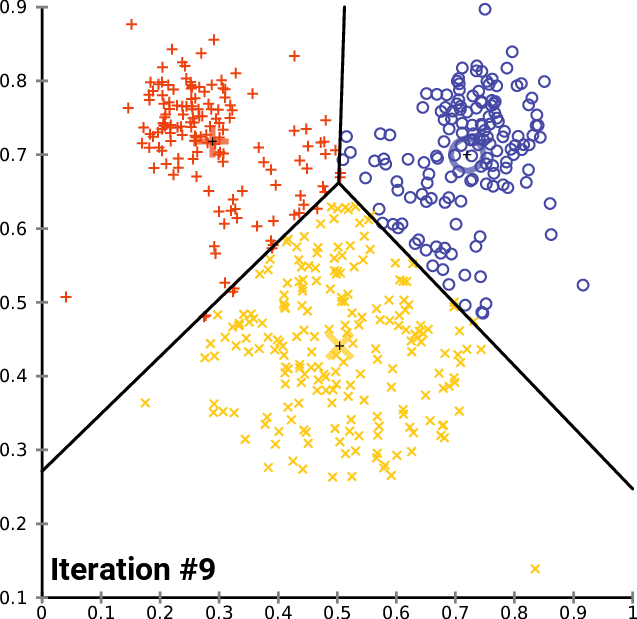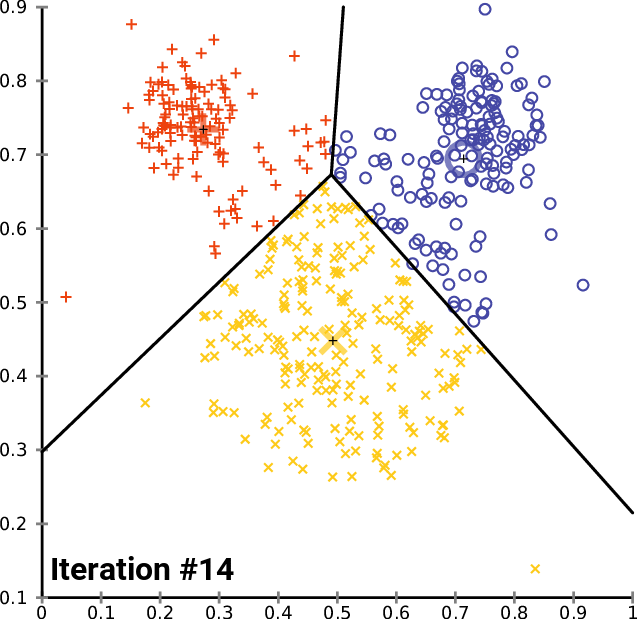
Example: Clustering countries by health and income
- Health expectancy in years vs. GDP per capita and population for 187 countries (as of 2015)
- Data from Gapminder
import altair as alt
from vega_datasets import data as vega_dataRead the data from a URL:
gapminder = pd.read_csv(vega_data.gapminder_health_income.url)
gapminder.head()| country | income | health | population | |
|---|---|---|---|---|
| 0 | Afghanistan | 1925 | 57.63 | 32526562 |
| 1 | Albania | 10620 | 76.00 | 2896679 |
| 2 | Algeria | 13434 | 76.50 | 39666519 |
| 3 | Andorra | 46577 | 84.10 | 70473 |
| 4 | Angola | 7615 | 61.00 | 25021974 |
Plot it with altair:
(
alt.Chart(gapminder)
.mark_circle()
.encode(
alt.X("income:Q", scale=alt.Scale(type="log")),
alt.Y("health:Q", scale=alt.Scale(zero=False)),
size="population:Q",
tooltip=list(gapminder.columns),
)
.properties(width=800, height=600)
.interactive()
)K-Means with scikit-learn
from sklearn.cluster import KMeansLet’s start with 5 clusters
KMeans?Init signature: KMeans( n_clusters=8, *, init='k-means++', n_init='warn', max_iter=300, tol=0.0001, verbose=0, random_state=None, copy_x=True, algorithm='lloyd', ) Docstring: K-Means clustering. Read more in the :ref:`User Guide <k_means>`. Parameters ---------- n_clusters : int, default=8 The number of clusters to form as well as the number of centroids to generate. init : {'k-means++', 'random'}, callable or array-like of shape (n_clusters, n_features), default='k-means++' Method for initialization: 'k-means++' : selects initial cluster centroids using sampling based on an empirical probability distribution of the points' contribution to the overall inertia. This technique speeds up convergence. The algorithm implemented is "greedy k-means++". It differs from the vanilla k-means++ by making several trials at each sampling step and choosing the best centroid among them. 'random': choose `n_clusters` observations (rows) at random from data for the initial centroids. If an array is passed, it should be of shape (n_clusters, n_features) and gives the initial centers. If a callable is passed, it should take arguments X, n_clusters and a random state and return an initialization. n_init : 'auto' or int, default=10 Number of times the k-means algorithm is run with different centroid seeds. The final results is the best output of `n_init` consecutive runs in terms of inertia. Several runs are recommended for sparse high-dimensional problems (see :ref:`kmeans_sparse_high_dim`). When `n_init='auto'`, the number of runs depends on the value of init: 10 if using `init='random'` or `init` is a callable; 1 if using `init='k-means++'` or `init` is an array-like. .. versionadded:: 1.2 Added 'auto' option for `n_init`. .. versionchanged:: 1.4 Default value for `n_init` will change from 10 to `'auto'` in version 1.4. max_iter : int, default=300 Maximum number of iterations of the k-means algorithm for a single run. tol : float, default=1e-4 Relative tolerance with regards to Frobenius norm of the difference in the cluster centers of two consecutive iterations to declare convergence. verbose : int, default=0 Verbosity mode. random_state : int, RandomState instance or None, default=None Determines random number generation for centroid initialization. Use an int to make the randomness deterministic. See :term:`Glossary <random_state>`. copy_x : bool, default=True When pre-computing distances it is more numerically accurate to center the data first. If copy_x is True (default), then the original data is not modified. If False, the original data is modified, and put back before the function returns, but small numerical differences may be introduced by subtracting and then adding the data mean. Note that if the original data is not C-contiguous, a copy will be made even if copy_x is False. If the original data is sparse, but not in CSR format, a copy will be made even if copy_x is False. algorithm : {"lloyd", "elkan", "auto", "full"}, default="lloyd" K-means algorithm to use. The classical EM-style algorithm is `"lloyd"`. The `"elkan"` variation can be more efficient on some datasets with well-defined clusters, by using the triangle inequality. However it's more memory intensive due to the allocation of an extra array of shape `(n_samples, n_clusters)`. `"auto"` and `"full"` are deprecated and they will be removed in Scikit-Learn 1.3. They are both aliases for `"lloyd"`. .. versionchanged:: 0.18 Added Elkan algorithm .. versionchanged:: 1.1 Renamed "full" to "lloyd", and deprecated "auto" and "full". Changed "auto" to use "lloyd" instead of "elkan". Attributes ---------- cluster_centers_ : ndarray of shape (n_clusters, n_features) Coordinates of cluster centers. If the algorithm stops before fully converging (see ``tol`` and ``max_iter``), these will not be consistent with ``labels_``. labels_ : ndarray of shape (n_samples,) Labels of each point inertia_ : float Sum of squared distances of samples to their closest cluster center, weighted by the sample weights if provided. n_iter_ : int Number of iterations run. n_features_in_ : int Number of features seen during :term:`fit`. .. versionadded:: 0.24 feature_names_in_ : ndarray of shape (`n_features_in_`,) Names of features seen during :term:`fit`. Defined only when `X` has feature names that are all strings. .. versionadded:: 1.0 See Also -------- MiniBatchKMeans : Alternative online implementation that does incremental updates of the centers positions using mini-batches. For large scale learning (say n_samples > 10k) MiniBatchKMeans is probably much faster than the default batch implementation. Notes ----- The k-means problem is solved using either Lloyd's or Elkan's algorithm. The average complexity is given by O(k n T), where n is the number of samples and T is the number of iteration. The worst case complexity is given by O(n^(k+2/p)) with n = n_samples, p = n_features. Refer to :doi:`"How slow is the k-means method?" D. Arthur and S. Vassilvitskii - SoCG2006.<10.1145/1137856.1137880>` for more details. In practice, the k-means algorithm is very fast (one of the fastest clustering algorithms available), but it falls in local minima. That's why it can be useful to restart it several times. If the algorithm stops before fully converging (because of ``tol`` or ``max_iter``), ``labels_`` and ``cluster_centers_`` will not be consistent, i.e. the ``cluster_centers_`` will not be the means of the points in each cluster. Also, the estimator will reassign ``labels_`` after the last iteration to make ``labels_`` consistent with ``predict`` on the training set. Examples -------- >>> from sklearn.cluster import KMeans >>> import numpy as np >>> X = np.array([[1, 2], [1, 4], [1, 0], ... [10, 2], [10, 4], [10, 0]]) >>> kmeans = KMeans(n_clusters=2, random_state=0, n_init="auto").fit(X) >>> kmeans.labels_ array([1, 1, 1, 0, 0, 0], dtype=int32) >>> kmeans.predict([[0, 0], [12, 3]]) array([1, 0], dtype=int32) >>> kmeans.cluster_centers_ array([[10., 2.], [ 1., 2.]]) File: ~/mambaforge/envs/musa-550-fall-2023/lib/python3.10/site-packages/sklearn/cluster/_kmeans.py Type: ABCMeta Subclasses:
kmeans = KMeans(n_clusters=5, n_init=10)Lot’s of optional parameters, but n_clusters is the most important:
Let’s fit just income first
Use the fit() function
kmeans.fit(gapminder[['income']]);Extract the cluster labels
Use the labels_ attribute
gapminder['label'] = kmeans.labels_How big are our clusters?
gapminder.groupby('label').size()label
0 106
1 6
2 50
3 1
4 24
dtype: int64Plot it again, coloring by our labels
(
alt.Chart(gapminder)
.mark_circle()
.encode(
alt.X("income:Q", scale=alt.Scale(type="log")),
alt.Y("health:Q", scale=alt.Scale(zero=False)),
size="population:Q",
color=alt.Color("label:N", scale=alt.Scale(scheme="dark2")),
tooltip=list(gapminder.columns),
)
.properties(width=800, height=600)
.interactive()
)Calculate average income by group
gapminder.groupby("label")['income'].mean().sort_values()label
0 5279.830189
2 21040.820000
4 42835.500000
1 74966.166667
3 132877.000000
Name: income, dtype: float64Data is nicely partitioned into income levels
How about health, income, and population?
# Fit all three columns
kmeans.fit(gapminder[['income', 'health', 'population']])
# Extract the labels
gapminder['label'] = kmeans.labels_gapminder| country | income | health | population | label | |
|---|---|---|---|---|---|
| 0 | Afghanistan | 1925 | 57.63 | 32526562 | 2 |
| 1 | Albania | 10620 | 76.00 | 2896679 | 2 |
| 2 | Algeria | 13434 | 76.50 | 39666519 | 0 |
| 3 | Andorra | 46577 | 84.10 | 70473 | 2 |
| 4 | Angola | 7615 | 61.00 | 25021974 | 2 |
| ... | ... | ... | ... | ... | ... |
| 182 | Vietnam | 5623 | 76.50 | 93447601 | 0 |
| 183 | West Bank and Gaza | 4319 | 75.20 | 4668466 | 2 |
| 184 | Yemen | 3887 | 67.60 | 26832215 | 2 |
| 185 | Zambia | 4034 | 58.96 | 16211767 | 2 |
| 186 | Zimbabwe | 1801 | 60.01 | 15602751 | 2 |
187 rows × 5 columns
(
alt.Chart(gapminder)
.mark_circle()
.encode(
alt.X("income:Q", scale=alt.Scale(type="log")),
alt.Y("health:Q", scale=alt.Scale(zero=False)),
size="population:Q",
color=alt.Color("label:N", scale=alt.Scale(scheme="dark2")),
tooltip=list(gapminder.columns),
)
.properties(width=800, height=600)
.interactive()
)It….didn’t work that well
What’s wrong?
K-means is distance-based, but our features have wildly different distance scales
scikit-learn to the rescue: pre-processing
- Scikit-learn has a utility to normalize features with an average of zero and a variance of 1
- Use the
StandardScalerclass
from sklearn.preprocessing import MinMaxScaler, RobustScalerfrom sklearn.preprocessing import StandardScalerscaler = StandardScaler()Use the fit_transform() function to scale your features
gapminder_scaled = scaler.fit_transform(gapminder[['income', 'health', 'population']])Important: The fit_transform() function converts the DataFrame to a numpy array:
# fit_transform() converts the data into a numpy array
gapminder_scaled[:5]array([[-0.79481258, -1.8171424 , -0.04592039],
[-0.34333373, 0.55986273, -0.25325837],
[-0.1972197 , 0.62456075, 0.00404216],
[ 1.52369617, 1.6079706 , -0.27303503],
[-0.49936524, -1.38107777, -0.09843447]])# mean of zero
gapminder_scaled.mean(axis=0)array([ 8.07434927e-17, -1.70511258e-15, -1.89984689e-17])# variance of one
gapminder_scaled.std(axis=0)array([1., 1., 1.])Now fit the scaled features
# Perform the fit
kmeans.fit(gapminder_scaled)
# Extract the labels
gapminder['label'] = kmeans.labels_(
alt.Chart(gapminder)
.mark_circle()
.encode(
alt.X("income:Q", scale=alt.Scale(type="log")),
alt.Y("health:Q", scale=alt.Scale(zero=False)),
size="population:Q",
color=alt.Color("label:N", scale=alt.Scale(scheme="dark2")),
tooltip=list(gapminder.columns),
)
.properties(width=800, height=600)
.interactive()
)# Number of countries per cluster
gapminder.groupby("label").size()label
0 62
1 85
2 5
3 2
4 33
dtype: int64# Average population per cluster
gapminder.groupby("label")['population'].mean().sort_values() / 1e6label
2 2.544302
0 21.191274
1 26.478761
4 31.674060
3 1343.549735
Name: population, dtype: float64# Average life expectancy per cluster
gapminder.groupby("label")['health'].mean().sort_values()label
0 62.342097
3 71.850000
1 74.376353
4 80.830303
2 80.920000
Name: health, dtype: float64# Average income per cluster
gapminder.groupby("label")['income'].mean().sort_values() / 1e3label
0 4.136016
3 9.618500
1 13.347376
4 41.048818
2 91.524200
Name: income, dtype: float64gapminder.loc[gapminder['label']==4]| country | income | health | population | label | |
|---|---|---|---|---|---|
| 3 | Andorra | 46577 | 84.1 | 70473 | 4 |
| 8 | Australia | 44056 | 81.8 | 23968973 | 4 |
| 9 | Austria | 44401 | 81.0 | 8544586 | 4 |
| 12 | Bahrain | 44138 | 79.2 | 1377237 | 4 |
| 16 | Belgium | 41240 | 80.4 | 11299192 | 4 |
| 30 | Canada | 43294 | 81.7 | 35939927 | 4 |
| 44 | Cyprus | 29797 | 82.6 | 1165300 | 4 |
| 45 | Czech Republic | 29437 | 78.6 | 10543186 | 4 |
| 46 | Denmark | 43495 | 80.1 | 5669081 | 4 |
| 58 | Finland | 38923 | 80.8 | 5503457 | 4 |
| 59 | France | 37599 | 81.9 | 64395345 | 4 |
| 63 | Germany | 44053 | 81.1 | 80688545 | 4 |
| 65 | Greece | 25430 | 79.8 | 10954617 | 4 |
| 74 | Iceland | 42182 | 82.8 | 329425 | 4 |
| 79 | Ireland | 47758 | 80.4 | 4688465 | 4 |
| 80 | Israel | 31590 | 82.4 | 8064036 | 4 |
| 81 | Italy | 33297 | 82.1 | 59797685 | 4 |
| 83 | Japan | 36162 | 83.5 | 126573481 | 4 |
| 104 | Malta | 30265 | 82.1 | 418670 | 4 |
| 118 | Netherlands | 45784 | 80.6 | 16924929 | 4 |
| 119 | New Zealand | 34186 | 80.6 | 4528526 | 4 |
| 124 | Norway | 64304 | 81.6 | 5210967 | 4 |
| 125 | Oman | 48226 | 75.7 | 4490541 | 4 |
| 133 | Portugal | 26437 | 79.8 | 10349803 | 4 |
| 140 | Saudi Arabia | 52469 | 78.1 | 31540372 | 4 |
| 147 | Slovenia | 28550 | 80.2 | 2067526 | 4 |
| 151 | South Korea | 34644 | 80.7 | 50293439 | 4 |
| 153 | Spain | 32979 | 81.7 | 46121699 | 4 |
| 160 | Sweden | 44892 | 82.0 | 9779426 | 4 |
| 161 | Switzerland | 56118 | 82.9 | 8298663 | 4 |
| 175 | United Arab Emirates | 60749 | 76.6 | 9156963 | 4 |
| 176 | United Kingdom | 38225 | 81.4 | 64715810 | 4 |
| 177 | United States | 53354 | 79.1 | 321773631 | 4 |
kmeans.inertia_80.72009578392847gapminder.loc[gapminder['label']==4]| country | income | health | population | label | |
|---|---|---|---|---|---|
| 3 | Andorra | 46577 | 84.1 | 70473 | 4 |
| 8 | Australia | 44056 | 81.8 | 23968973 | 4 |
| 9 | Austria | 44401 | 81.0 | 8544586 | 4 |
| 12 | Bahrain | 44138 | 79.2 | 1377237 | 4 |
| 16 | Belgium | 41240 | 80.4 | 11299192 | 4 |
| 30 | Canada | 43294 | 81.7 | 35939927 | 4 |
| 44 | Cyprus | 29797 | 82.6 | 1165300 | 4 |
| 45 | Czech Republic | 29437 | 78.6 | 10543186 | 4 |
| 46 | Denmark | 43495 | 80.1 | 5669081 | 4 |
| 58 | Finland | 38923 | 80.8 | 5503457 | 4 |
| 59 | France | 37599 | 81.9 | 64395345 | 4 |
| 63 | Germany | 44053 | 81.1 | 80688545 | 4 |
| 65 | Greece | 25430 | 79.8 | 10954617 | 4 |
| 74 | Iceland | 42182 | 82.8 | 329425 | 4 |
| 79 | Ireland | 47758 | 80.4 | 4688465 | 4 |
| 80 | Israel | 31590 | 82.4 | 8064036 | 4 |
| 81 | Italy | 33297 | 82.1 | 59797685 | 4 |
| 83 | Japan | 36162 | 83.5 | 126573481 | 4 |
| 104 | Malta | 30265 | 82.1 | 418670 | 4 |
| 118 | Netherlands | 45784 | 80.6 | 16924929 | 4 |
| 119 | New Zealand | 34186 | 80.6 | 4528526 | 4 |
| 124 | Norway | 64304 | 81.6 | 5210967 | 4 |
| 125 | Oman | 48226 | 75.7 | 4490541 | 4 |
| 133 | Portugal | 26437 | 79.8 | 10349803 | 4 |
| 140 | Saudi Arabia | 52469 | 78.1 | 31540372 | 4 |
| 147 | Slovenia | 28550 | 80.2 | 2067526 | 4 |
| 151 | South Korea | 34644 | 80.7 | 50293439 | 4 |
| 153 | Spain | 32979 | 81.7 | 46121699 | 4 |
| 160 | Sweden | 44892 | 82.0 | 9779426 | 4 |
| 161 | Switzerland | 56118 | 82.9 | 8298663 | 4 |
| 175 | United Arab Emirates | 60749 | 76.6 | 9156963 | 4 |
| 176 | United Kingdom | 38225 | 81.4 | 64715810 | 4 |
| 177 | United States | 53354 | 79.1 | 321773631 | 4 |
Exercise: Clustering neighborhoods by Airbnb stats
I’ve extracted neighborhood Airbnb statistics for Philadelphia neighborhoods from Tom Slee’s website.
The data includes average price per person, overall satisfaction, and number of listings.
Original research study: How Airbnb’s Data Hid the Facts in New York City
Step 1: Load the data with pandas
The data is available in CSV format (“philly_airbnb_by_neighborhoods.csv”) in the “data/” folder of the repository.
airbnb = pd.read_csv("data/philly_airbnb_by_neighborhoods.csv")
airbnb.head()| neighborhood | price_per_person | overall_satisfaction | N | |
|---|---|---|---|---|
| 0 | ALLEGHENY_WEST | 120.791667 | 4.666667 | 23 |
| 1 | BELLA_VISTA | 87.407920 | 3.158333 | 204 |
| 2 | BELMONT | 69.425000 | 3.250000 | 11 |
| 3 | BREWERYTOWN | 71.788188 | 1.943182 | 142 |
| 4 | BUSTLETON | 55.833333 | 1.250000 | 19 |
Step 2: Perform the K-Means fit
- Use our three features:
price_per_person,overall_satisfaction,N - I used 5 clusters, but you are welcome to experiment with different values! We will discuss what the optimal number is after we go through the solutions!
- Scaling the features is recommended, but if the scales aren’t too different, so probably isn’t necessary in this case
# Initialize the Kmeans object
kmeans = KMeans(n_clusters=5, random_state=42, n_init=10)Pre-process the data:
# Scale the data features we want
scaler = StandardScaler()
scaled_airbnb_data = scaler.fit_transform(airbnb[['price_per_person', 'overall_satisfaction', 'N']])scaled_airbnb_data[:5]array([[ 0.79001045, 1.74917003, -0.52348653],
[ 0.0714296 , 0.41880366, 0.40700931],
[-0.31565045, 0.49965466, -0.58517687],
[-0.26478314, -0.65297315, 0.08827593],
[-0.60820933, -1.26436704, -0.54404998]])# Run the fit! This adds the ".labels_" attribute
kmeans.fit(scaled_airbnb_data);kmeans.labels_array([0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 0, 4, 4, 0,
1, 4, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 4, 0, 0, 1, 0, 0, 0, 0, 1,
0, 1, 0, 4, 0, 0, 0, 0, 1, 0, 0, 0, 0, 4, 0, 0, 4, 0, 0, 1, 1, 1,
0, 0, 0, 1, 4, 0, 4, 1, 0, 3, 0, 0, 2, 4, 1, 0, 4, 4, 0, 1, 1, 0,
4, 0, 0, 4, 0, 1, 0, 0, 0, 0, 0, 1, 1, 0, 0, 1, 1], dtype=int32)# Save the cluster labels
airbnb["label"] = kmeans.labels_# New column "label"!
airbnb.head()| neighborhood | price_per_person | overall_satisfaction | N | label | |
|---|---|---|---|---|---|
| 0 | ALLEGHENY_WEST | 120.791667 | 4.666667 | 23 | 0 |
| 1 | BELLA_VISTA | 87.407920 | 3.158333 | 204 | 0 |
| 2 | BELMONT | 69.425000 | 3.250000 | 11 | 0 |
| 3 | BREWERYTOWN | 71.788188 | 1.943182 | 142 | 0 |
| 4 | BUSTLETON | 55.833333 | 1.250000 | 19 | 1 |
Step 3: Calculate average features per cluster
To gain some insight into our clusters, after calculating the K-Means labels: - group by the label column - calculate the mean() of each of our features - calculate the number of neighborhoods per cluster
airbnb.groupby('label', as_index=False).size()| label | size | |
|---|---|---|
| 0 | 0 | 69 |
| 1 | 1 | 19 |
| 2 | 2 | 1 |
| 3 | 3 | 1 |
| 4 | 4 | 15 |
airbnb.groupby("label", as_index=False)[
["price_per_person", "overall_satisfaction", "N"]
].mean().sort_values(by="price_per_person")| label | price_per_person | overall_satisfaction | N | |
|---|---|---|---|---|
| 0 | 0 | 73.199020 | 3.137213 | 76.550725 |
| 1 | 1 | 79.250011 | 0.697461 | 23.473684 |
| 4 | 4 | 116.601261 | 2.936508 | 389.933333 |
| 3 | 3 | 136.263996 | 3.000924 | 1499.000000 |
| 2 | 2 | 387.626984 | 5.000000 | 31.000000 |
airbnb.loc[airbnb['label'] == 2]| neighborhood | price_per_person | overall_satisfaction | N | label | |
|---|---|---|---|---|---|
| 78 | SHARSWOOD | 387.626984 | 5.0 | 31 | 2 |
airbnb.loc[airbnb['label'] == 3]| neighborhood | price_per_person | overall_satisfaction | N | label | |
|---|---|---|---|---|---|
| 75 | RITTENHOUSE | 136.263996 | 3.000924 | 1499 | 3 |
airbnb.loc[airbnb['label'] == 4]| neighborhood | price_per_person | overall_satisfaction | N | label | |
|---|---|---|---|---|---|
| 16 | EAST_PARK | 193.388889 | 2.714286 | 42 | 4 |
| 19 | FAIRMOUNT | 144.764110 | 2.903614 | 463 | 4 |
| 20 | FISHTOWN | 59.283468 | 2.963816 | 477 | 4 |
| 23 | FRANCISVILLE | 124.795795 | 3.164062 | 300 | 4 |
| 35 | GRADUATE_HOSPITAL | 106.420417 | 3.180791 | 649 | 4 |
| 47 | LOGAN_SQUARE | 145.439414 | 3.139241 | 510 | 4 |
| 57 | NORTHERN_LIBERTIES | 145.004866 | 3.095506 | 367 | 4 |
| 60 | OLD_CITY | 111.708084 | 2.756637 | 352 | 4 |
| 70 | POINT_BREEZE | 63.801072 | 2.759542 | 435 | 4 |
| 72 | QUEEN_VILLAGE | 106.405744 | 3.125000 | 248 | 4 |
| 79 | SOCIETY_HILL | 133.598667 | 3.118421 | 165 | 4 |
| 82 | SPRING_GARDEN | 157.125692 | 3.454023 | 413 | 4 |
| 83 | SPRUCE_HILL | 48.095512 | 2.377358 | 399 | 4 |
| 88 | UNIVERSITY_CITY | 82.228062 | 2.231579 | 326 | 4 |
| 91 | WASHINGTON_SQUARE | 126.959118 | 3.063745 | 703 | 4 |
Step 4: Plot a choropleth, coloring neighborhoods by their cluster label
- Part 1: Load the Philadelphia neighborhoods available in the data directory:
./data/philly_neighborhoods.geojson
- Part 2: Merge the Airbnb data (with labels) and the neighborhood polygons
- Part 3: Use geopandas to plot the neighborhoods
- The
categorical=Trueandlegend=Truekeywords will be useful here
- The
hoods = gpd.read_file("./data/philly_neighborhoods.geojson")
hoods.head()| Name | geometry | |
|---|---|---|
| 0 | LAWNDALE | POLYGON ((-75.08616 40.05013, -75.08893 40.044... |
| 1 | ASTON_WOODBRIDGE | POLYGON ((-75.00860 40.05369, -75.00861 40.053... |
| 2 | CARROLL_PARK | POLYGON ((-75.22673 39.97720, -75.22022 39.974... |
| 3 | CHESTNUT_HILL | POLYGON ((-75.21278 40.08637, -75.21272 40.086... |
| 4 | BURNHOLME | POLYGON ((-75.08768 40.06861, -75.08758 40.068... |
airbnb.head()| neighborhood | price_per_person | overall_satisfaction | N | label | |
|---|---|---|---|---|---|
| 0 | ALLEGHENY_WEST | 120.791667 | 4.666667 | 23 | 0 |
| 1 | BELLA_VISTA | 87.407920 | 3.158333 | 204 | 0 |
| 2 | BELMONT | 69.425000 | 3.250000 | 11 | 0 |
| 3 | BREWERYTOWN | 71.788188 | 1.943182 | 142 | 0 |
| 4 | BUSTLETON | 55.833333 | 1.250000 | 19 | 1 |
# Do the merge
# Note: how='left' will keep ALL of the geometries, even if they don't have any AirBnB data
airbnb2 = hoods.merge(airbnb, left_on='Name', right_on='neighborhood', how='left')
# Some neigborhoods don't have listings and get assigned the NaN label
# Assign -1 to the neighborhoods without any listings instead
airbnb2['label'] = airbnb2['label'].fillna(-1)# plot the data
airbnb2 = airbnb2.to_crs(epsg=3857)
# setup the figure
f, ax = plt.subplots(figsize=(10, 8))
# plot, coloring by label column
# specify categorical data and add legend
airbnb2.plot(
column="label",
cmap="Dark2",
categorical=True,
legend=True,
edgecolor="k",
lw=0.5,
ax=ax,
)
ax.set_axis_off()
plt.axis("equal");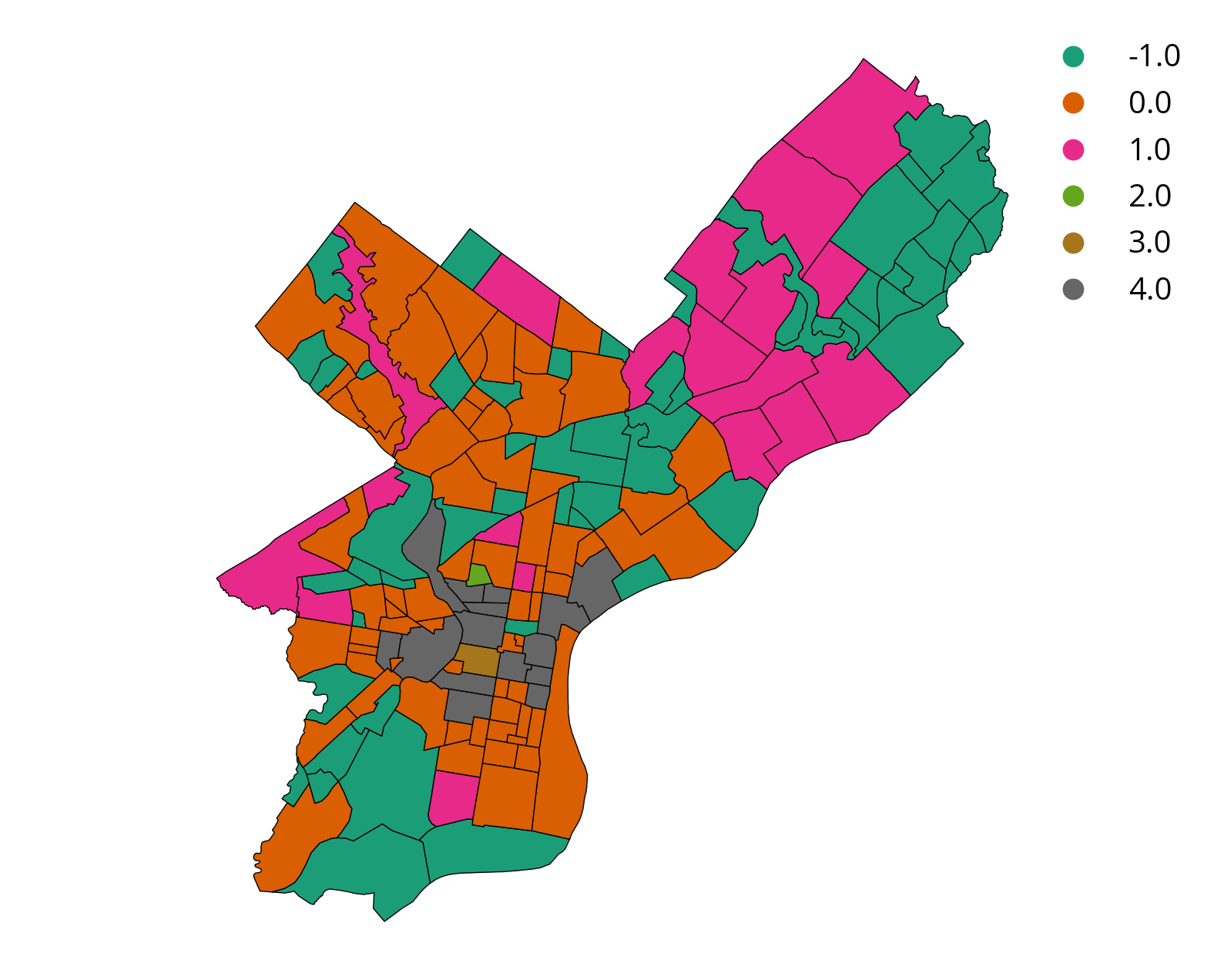
Step 5: Plot an interactive map
Use hvplot / geopandas to plot the clustering results with a tooltip for neighborhood name and tooltip.
airbnb2.explore(
column="label",
cmap="Dark2",
categorical=True,
legend=True,
tiles="CartoDB positron"
)Based on these results, where would you want to stay?
Group 0!
Why 5 groups?
Use the “Elbow” method…
# Number of clusters to try out
n_clusters = list(range(2, 10))
# Run kmeans for each value of k
inertias = []
for k in n_clusters:
# Initialize and run
kmeans = KMeans(n_clusters=k, n_init=10)
kmeans.fit(scaled_airbnb_data)
# Save the "inertia"
inertias.append(kmeans.inertia_)
# Plot it!
plt.plot(n_clusters, inertias, marker='o', ms=10, mfc='white', lw=4, mew=3);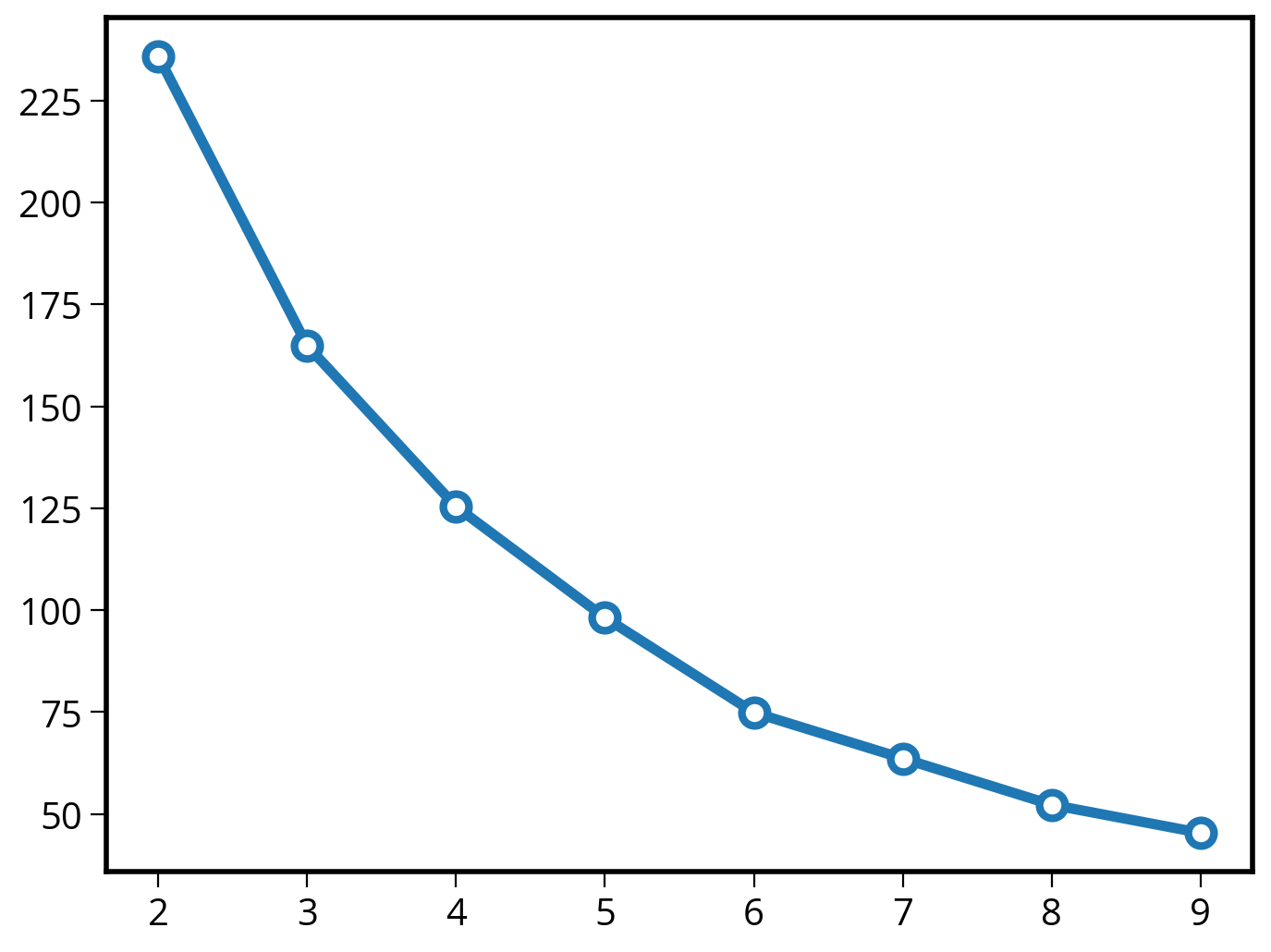
The kneed package
There is also a nice package called kneed that can determine the “knee” point quantitatively, using the kneedle algorithm.
n_clusters[2, 3, 4, 5, 6, 7, 8, 9]from kneed import KneeLocator
# Initialize the knee algorithm
kn = KneeLocator(n_clusters, inertias, curve='convex', direction='decreasing')
# Print out the knee
print(kn.knee)5That’s it!
More clustering on Wednesday!